Biology:Intro to gene expression central dogma)
How genes in DNA can provide instructions for proteins. The central dogma of molecular biology: DNA → RNA → protein.
Overview: Gene expression
DNA is the genetic material of all organisms on Earth. When DNA is transmitted from parents to children, it can determine some of the children's characteristics (such as their eye color or hair color). But how does the sequence of a DNA molecule actually affect a human or other organism's features? For example, how did the sequence of nucleotides (As, Ts, Cs, and Gs) in the DNA of Mendel's pea plants determine the color of their flowers?
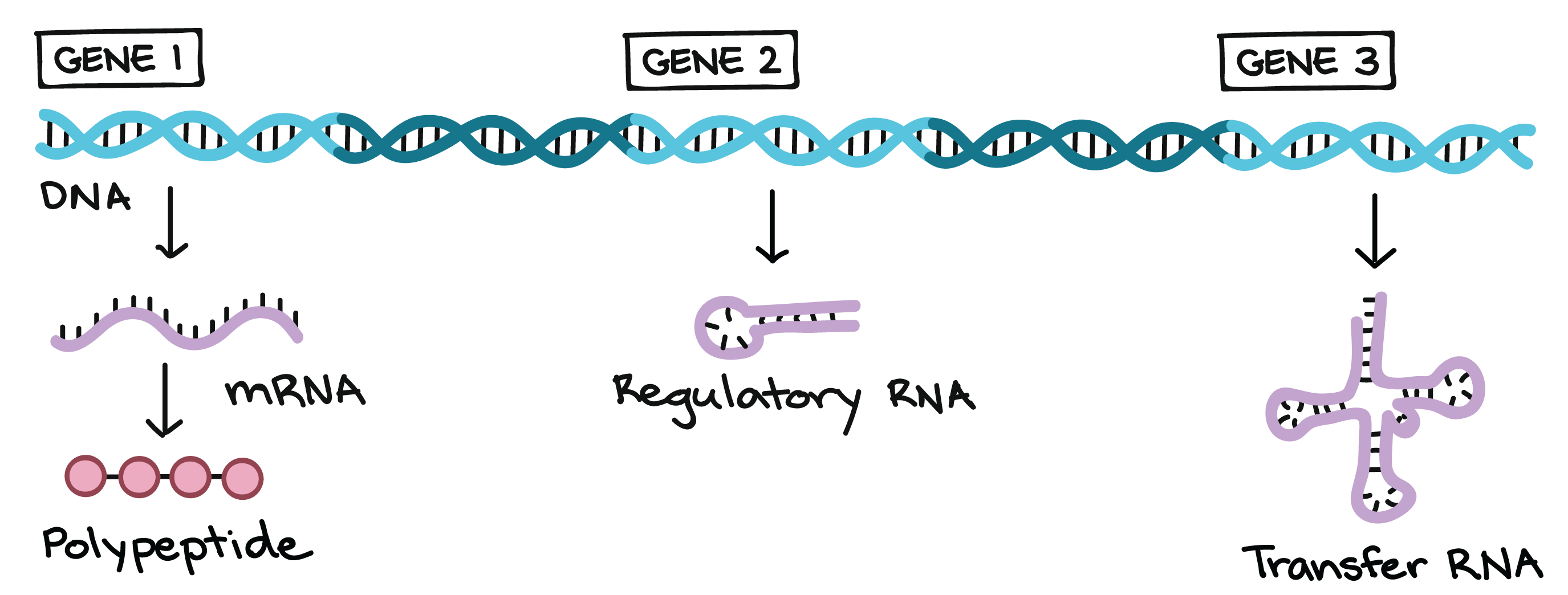
Genes specify functional products (such as proteins)
A DNA molecule isn't just a long, boring string of nucleotides. Instead, it's divided up into functional units called genes. Each gene provides instructions for a functional product, that is, a molecule needed to perform a job in the cell. In many cases, the functional product of a gene is a protein. For example, Mendel's flower color gene provides instructions for a protein that helps make colored molecules (pigments) in flower petals.
The functional products of most known genes are proteins, or, more accurately, polypeptides. Polypeptide is just another word for a chain of amino acids. Although many proteins consist of a single polypeptide, some are made up of multiple polypeptides. Genes that specify polypeptides are called protein-coding genes.
Not all genes specify polypeptides. Instead, some provide instructions to build functional RNA molecules, such as the transfer RNAs and ribosomal RNAsthat play roles in translation.
How does the DNA sequence of a gene specify a particular protein?
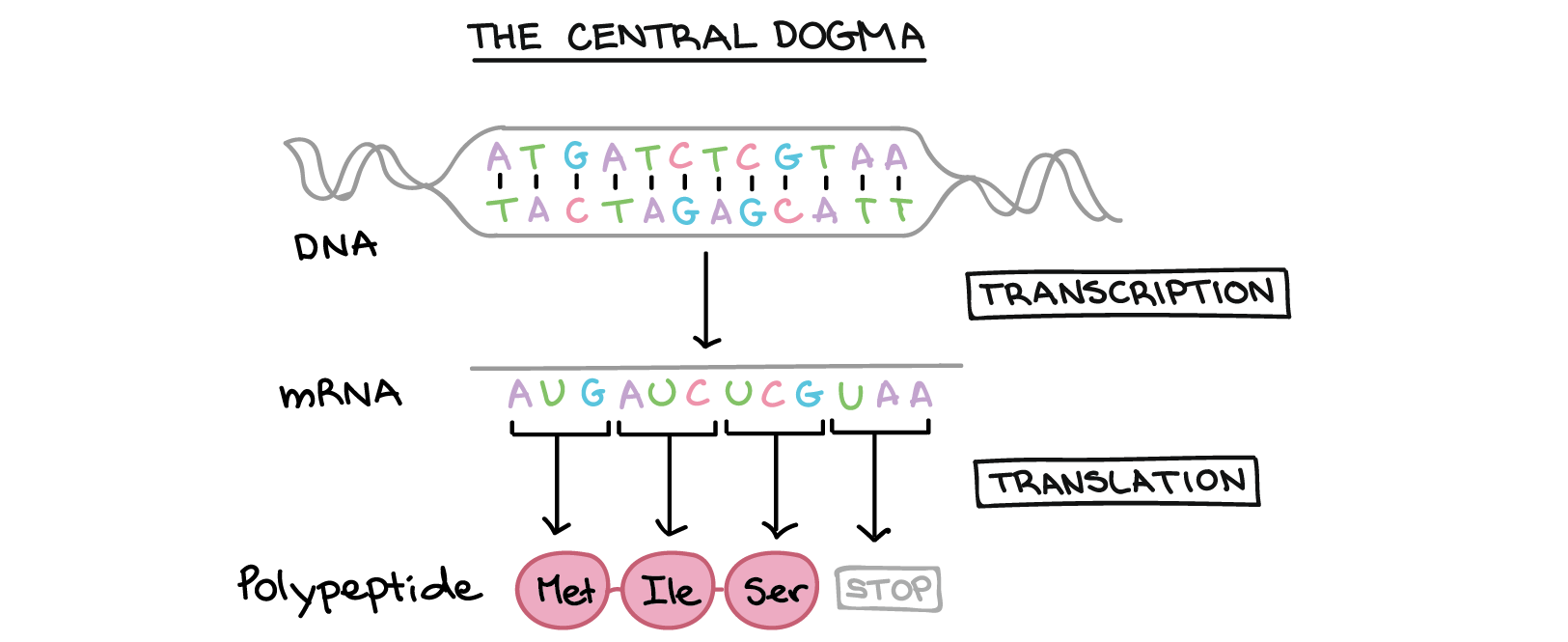
Many genes provide instructions for building polypeptides. How, exactly, does DNA direct the construction of a polypeptide? This process involves two major steps: transcription and translation.
- In transcription, the DNA sequence of a gene is copied to make an RNA molecule. This step is called transcription because it involves rewriting, or transcribing, the DNA sequence in a similar RNA "alphabet." In eukaryotes, the RNA molecule must undergo processing to become a mature messenger RNA (mRNA).
- In translation, the sequence of the mRNA is decoded to specify the amino acid sequence of a polypeptide. The name translation reflects that the nucleotide sequence of the mRNA sequence must be translated into the completely different "language" of amino acids.
Thus, during expression of a protein-coding gene, information flows from DNA right arrow RNA right arrow protein. This directional flow of information is known as the central dogma of molecular biology. Non-protein-coding genes (genes that specify functional RNAs) are still transcribed to produce an RNA, but this RNA is not translated into a polypeptide. For either type of gene, the process of going from DNA to a functional product is known as gene expression.
Transcription
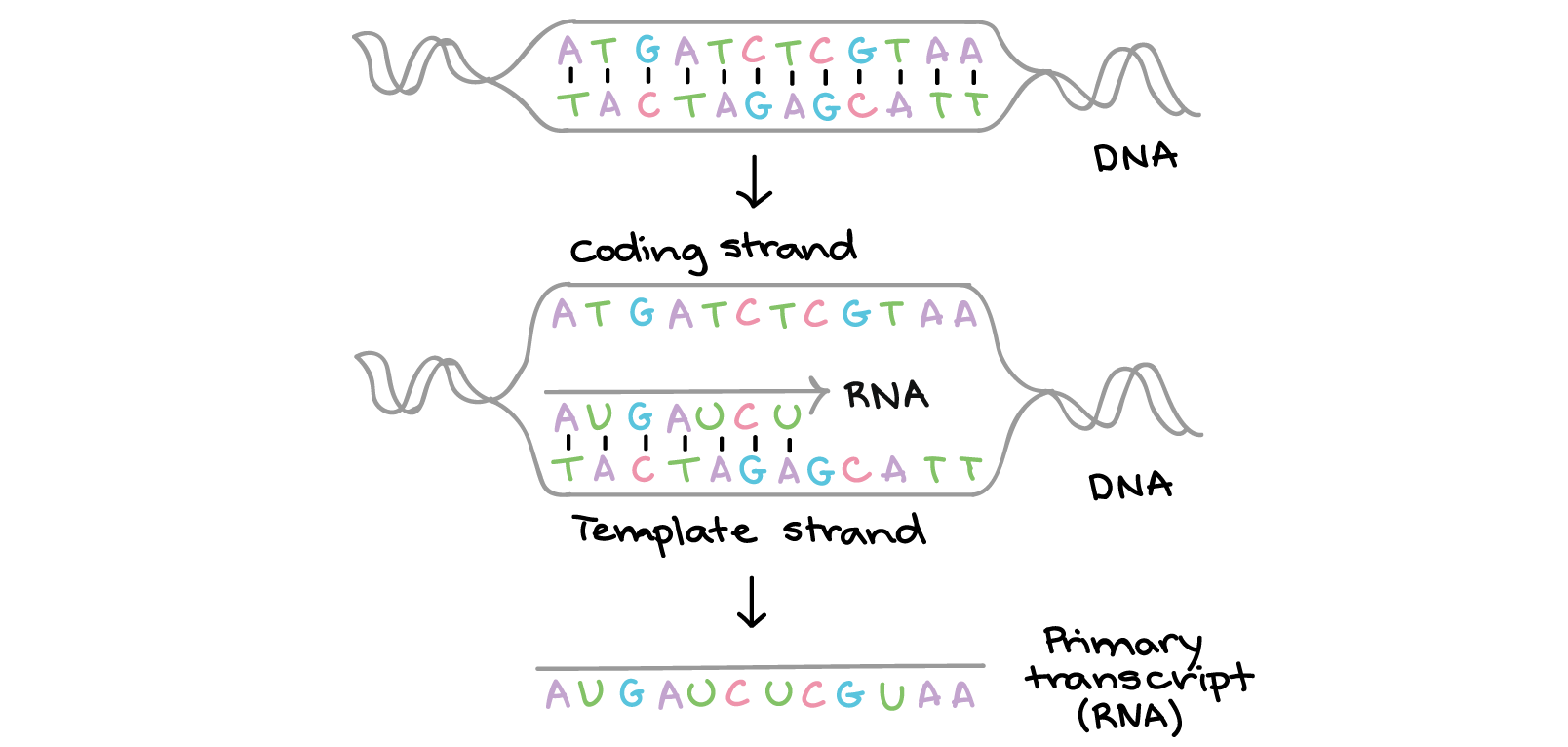
In transcription, one strand of the DNA that makes up a gene, called the non-coding strand, acts as a template for the synthesis of a matching (complementary) RNA strand by an enzyme called RNA polymerase. This RNA strand is the primary transcript.
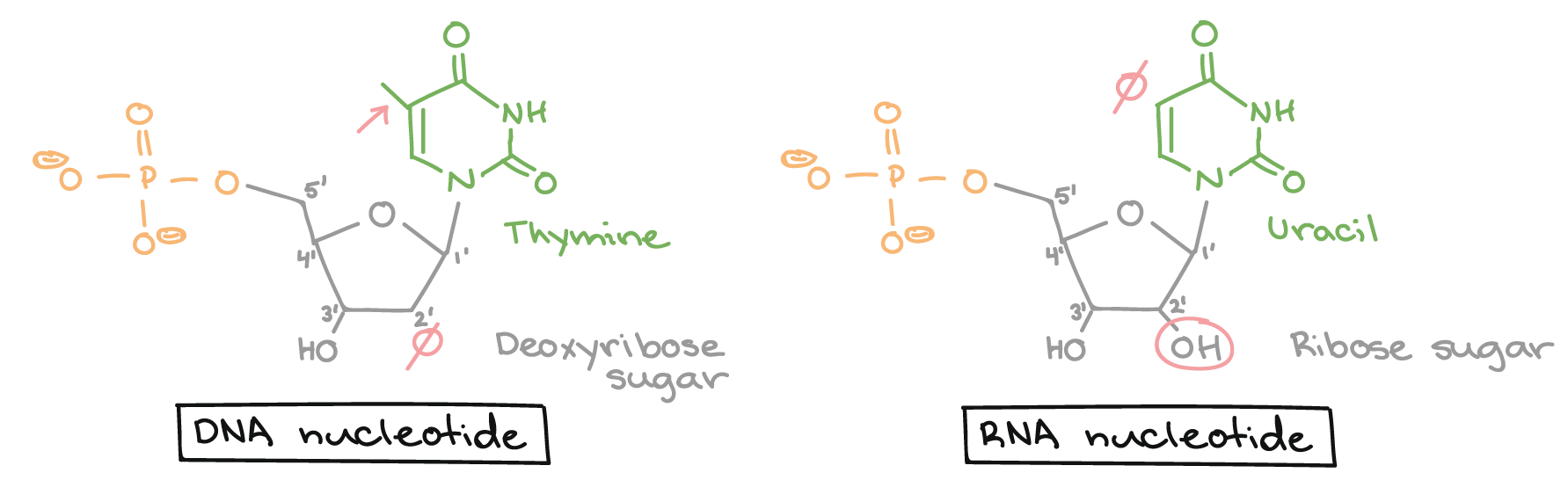
The primary transcript carries the same sequence information as the non-transcribed strand of DNA, sometimes called the coding strand. However, the primary transcript and the coding strand of DNA are not identical, thanks to some biochemical differences between DNA and RNA. One important difference is that RNA molecules do not include the base thymine (T). Instead, they have the similar base uracil (U). Like thymine, uracil pairs with adenine.
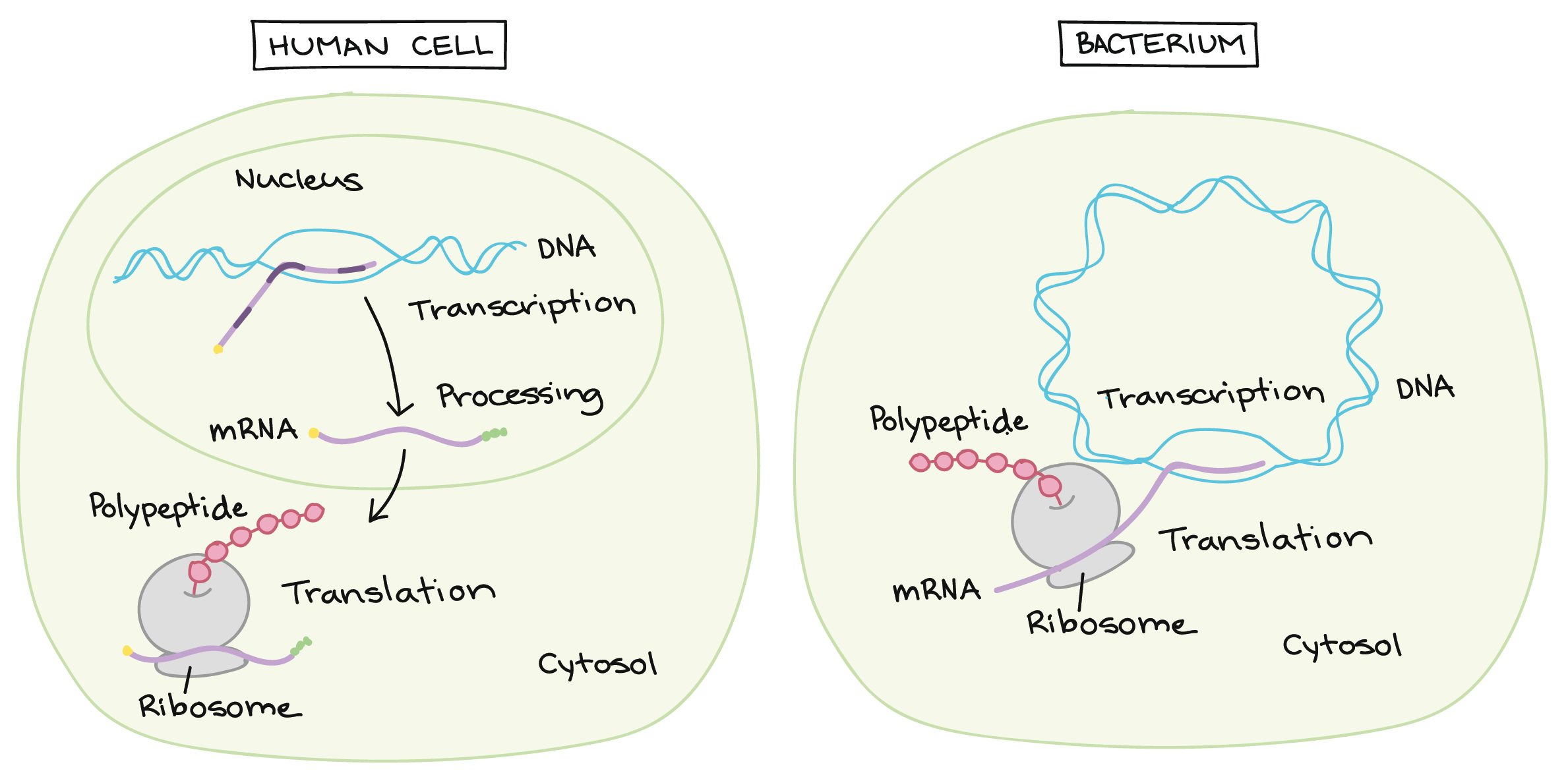
Transcription and RNA processing: Eukaryotes vs. bacteria
In bacteria, the primary RNA transcript can directly serve as a messenger RNA, or mRNA. Messenger RNAs get their name because they act as messengers between DNA and ribosomes. Ribosomes are RNA-and-protein structures in the cytosol where proteins are actually made.
In eukaryotes (such as humans), a primary transcript has to go through some extra processing steps in order to become a mature mRNA. During processing, caps are added to the ends of the RNA, and some pieces of it may be carefully removed in a process called splicing. These steps do not happen in bacteria.
The location of transcription is also different between prokaryotes and eukaryotes. Eukaryotic transcription takes place in the nucleus, where the DNA is stored, while protein synthesis takes place in the cytosol. Because of this, a eukaryotic mRNA must be exported from the nucleus before it can be translated into a polypeptide. Prokaryotic cells, on the other hand, don't have a nucleus, so they carry out both transcription and translation in the cytosol.
Translation
After transcription (and, in eukaryotes, after processing), an mRNA molecule is ready to direct protein synthesis. The process of using information in an mRNA to build a polypeptide is called translation.
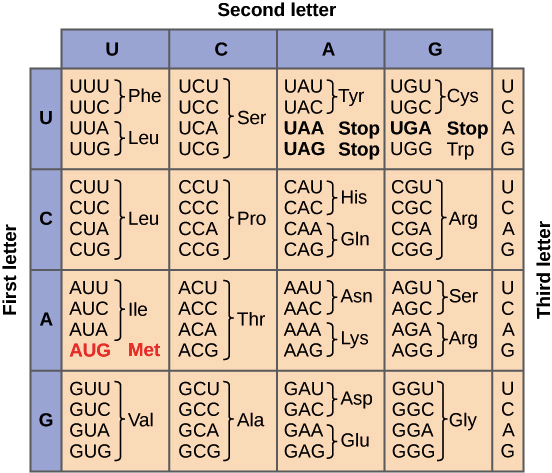
The genetic code
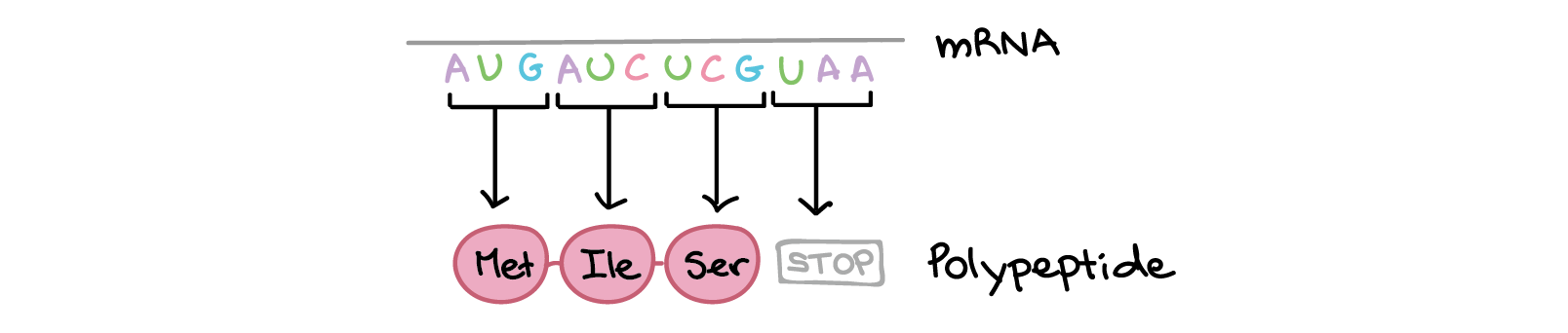
During translation, the nucleotide sequence of an mRNA is translated into the amino acid sequence of a polypeptide. Specifically, the nucleotides of the mRNA are read in triplets (groups of three) called codons. There are 61 codons that specify amino acids. One codon is a "start" codon that indicates where to start translation. The start codon specifies the amino acid methionine, so most polypeptides begin with this amino acid. Three other “stop” codons signal the end of a polypeptide. These relationships between codons and amino acids are called the genetic code.
Steps of translation
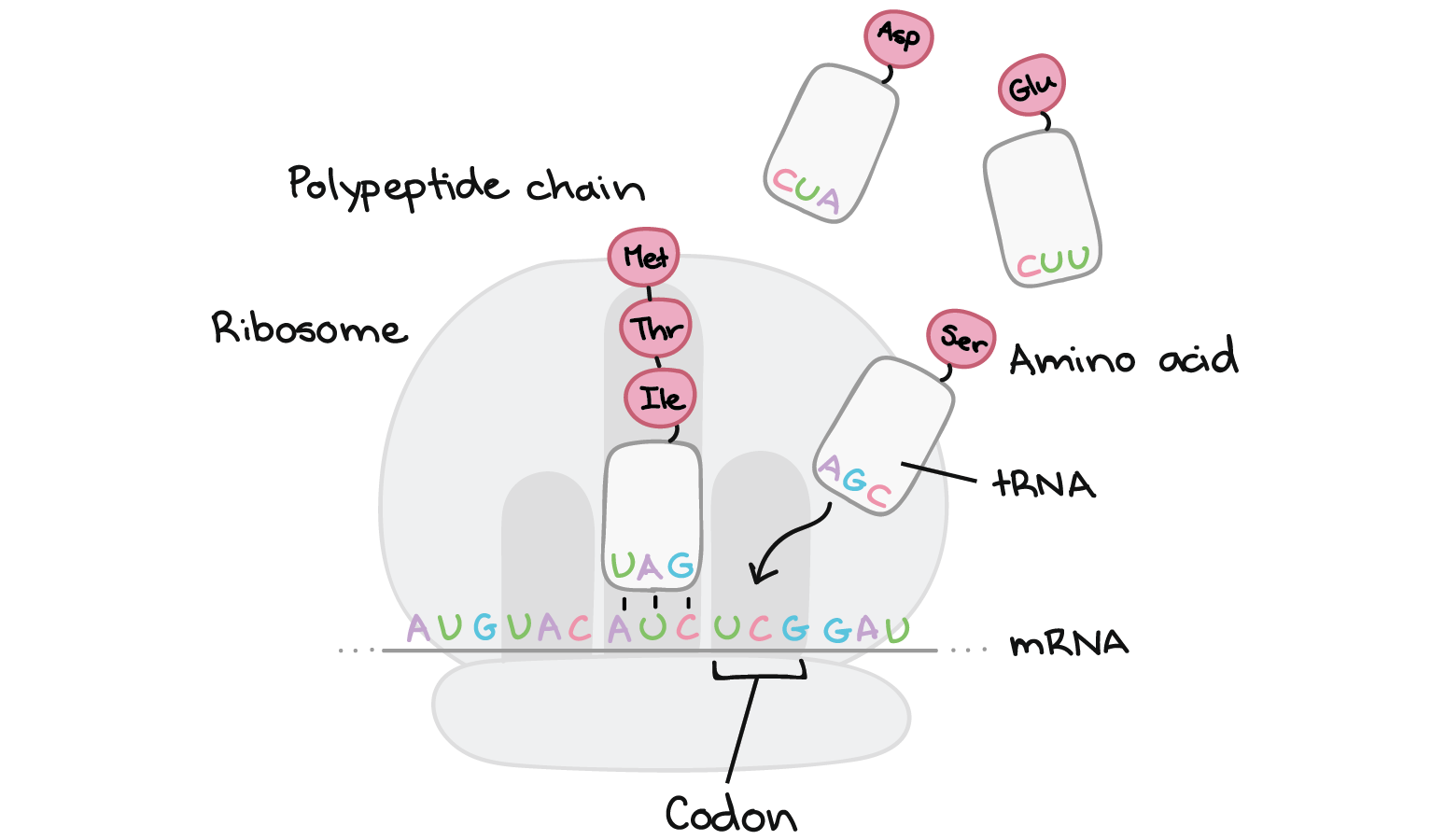
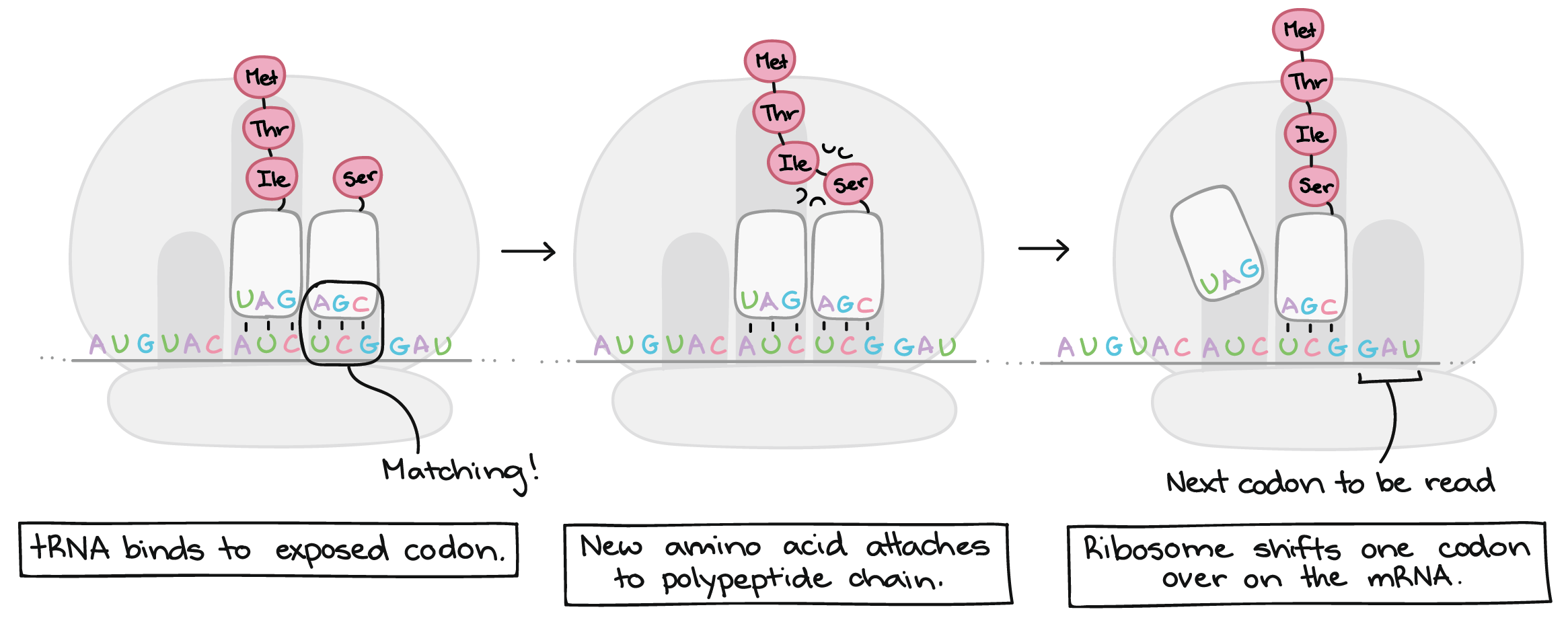
Translation takes place inside of structures known as ribosomes. Ribosomes are molecular machines whose job is to build polypeptides. Once a ribosome latches on to an mRNA and finds the "start" codon, it will travel rapidly down the mRNA, one codon at a time. As it goes, it will gradually build a chain of amino acids that exactly mirrors the sequence of codons in the mRNA.
How does the ribosome "know" which amino acid to add for each codon? As it turns out, this matching is not done by the ribosome itself. Instead, it depends on a group of specialized RNA molecules called transfer RNAS (tRNAs). Each tRNA has a three nucleotides sticking out at one end, which can recognize (base-pair with) just one or a few particular codons. At the other end, the tRNA carries an amino acid – specifically, the amino acid that matches those codons.
There are many tRNAs floating around in a cell, but only a tRNA that matches (base-pairs with) the codon that's currently being read can bind and deliver its amino acid cargo. Once a tRNA is snugly bound to its matching codon in the ribosome, its amino acid will be added the end of the polypeptide chain.
This process repeats many times, with the ribosome moving down the mRNA one codon at a time. A chain of amino acids is built up one by one, with an amino acid sequence that matches the sequence of codons found in the mRNA. Translation ends when the ribosome reaches a stop codon and releases the polypeptide.
What happens next?
Once the polypeptide is finished, it may be processed or modified, combine with other polypeptides, or be shipped to a specific destination inside or outside the cell. Ultimately, it will perform a specific job needed by the cell or organism – perhaps as a signaling molecule, structural element, or enzyme!
Summary:
- DNA is divided up into functional units called genes, which may specify polypeptides (proteins and protein subunits) or functional RNAs (such as tRNAs and rRNAs).
- Information from a gene is used to build a functional product in a process called gene expression.
- A gene that encodes a polypeptide is expressed in two steps. In this process, information flows from DNA right arrow RNA right arrow protein, a directional relationship known as the central dogma of molecular biology.
- Transcription: One strand of the gene's DNA is copied into RNA. In eukaryotes, the RNA transcript must undergo additional processing steps in order to become a mature messenger RNA (mRNA).
- Translation: The nucleotide sequence of the mRNA is decoded to specify the amino acid sequence of a polypeptide. This process occurs inside a ribosome and requires adapter molecules called tRNAs.
- During translation, the nucleotides of the mRNA are read in groups of three called codons. Each codon specifies a particular amino acid or a stop signal. This set of relationships is known as the genetic code.
- source:https://www.khanacademy.org/science/high-school-biology/hs-molecular-genetics/hs-rna-and-protein-synthesis/a/intro-to-gene-expression-central-dogma
Astronomy
Types and Classification of Galaxies
There are three main types of galaxies: Elliptical, Spiral, and Irregular. Two of these three types are further divided and classified into a system that is now known the tuning fork diagram. When Hubble first created this diagram, he believed that this was an evolutionary sequence as well as a classification.Elliptical Galaxies
| Elliptical galaxies are shaped like a spheriod, or elongated sphere. In the sky, where we can only see two of their three dimensions, these galaxies look like elliptical, or oval, shaped disks. The light is smooth, with the surface brightness decreasing as you go farther out from the center. Elliptical galaxies are given a classification that corresponds to their elongation from a perfect circle, otherwise known as their ellipticity. The larger the number, the more elliptical the galaxy is. So, for example a galaxy of classification of E0 appears to be perfectly circular, while a classification of E7 is very flattened. The elliptical scale varies from E0 to E7. Elliptical galaxies have no particular axis of rotation. |  Elliptical galaxy M87 Elliptical galaxy M87 |
Spiral Galaxies
 Spiral galaxy M100 Spiral galaxy M100 | Spiral galaxies have three main components: a bulge, disk, and halo (see right). The bulge is a spherical structure found in the center of the galaxy. This feature mostly contains older stars. The disk is made up of dust, gas, and younger stars. The disk forms arm structures. Our Sun is located in an arm of our galaxy, the Milky Way. The halo of a galaxy is a loose, spherical structure located around the bulge and some of the disk. The halo contains old clusters of stars, known as globular clusters. |  |
| Spiral galaxies are classified into two groups, ordinary and barred. The ordinary group is designated by S or SA, and the barred group by SB. In normal spirals (as seen at above left) the arms originate directly from the nucleus, or bulge, where in the barred spirals (see right) there is a bar of material that runs through the nucleus that the arms emerge from. Both of these types are given a classification according to how tightly their arms are wound. The classifications are a, b, c, d ... with "a" having the tightest arms. In type "a", the arms are usually not well defined and form almost a circular pattern. Sometimes you will see the classification of a galaxy with two lower case letters. This means that the tightness of the spiral structure is halfway between those two letters. |  Spiral galaxy NGC 1365 Spiral galaxy NGC 1365 |
S0 Galaxies
| S0 galaxies are an intermediate type of galaxy between E7 and a "true" spiral Sa. They differ from ellipticals because they have a bulge and a thin disk, but are different from Sa because they have no spiral structure. S0 galaxies are also known as Lenticular galaxies. |
Irregular Galaxies
| Irregular galaxies have no regular or symmetrical structure. They are divided into two groups, Irr I and IrrII. Irr I type galaxies have HII regions, which are regions of elemental hydrogen gas, and many Population I stars, which are young hot stars. Irr II galaxies simply seem to have large amounts of dust that block most of the light from the stars. All this dust makes is almost impossible to see distinct stars in the galaxy. Source:http://hosting.astro.cornell.edu/academics/courses/astro201/galaxies/types.htm |  Large Magellanic Cloud Large Magellanic Cloud |

No comments:
Post a Comment